Project Information
- 170308 Klebsormidium flaccidum NIES-2285 was re-identified as Klebsormidium nitens NIES-2285, 2016 Jun
- 140528 Klebsormidium flaccidum genome V1.0 released
Sequence data have been deposited in GenBank/EMBL/DDBJ Accession number: DF236950-DF238763 (1814 entries)
BioProject ID: PRJDB718
Release Information
- 171026 V1.1 General feature format file was revised (add missing mRNA and exon features of 43 genes).
- 170308 Klebsormidium nitens NIES-2285 V 1.1 General feature format release (DOWNLOAD)
- 160619 Klebsormidium nitens transcripts V1.1 released (DOWNLOAD)
- 140528 Klebsormidium nitens genome V1.0 released (DOWNLOAD)
(K. flaccidum NIES-2285 was re-identified as K. nitens NIES-2285, 2016 Jun)
Introduction
- Klebsormidium nitens NIES-2145 The charophytic algae Klebsormidium nitens strain NIES-2285 (formerly identified as K. flaccidum) usually consist of multicellular and non-branching filaments without differentiated or specialised cells.
Each cell has a large chloroplast lying against cell wall (parietal chloroplast). pyrenoid surrounded by a few starch grains.
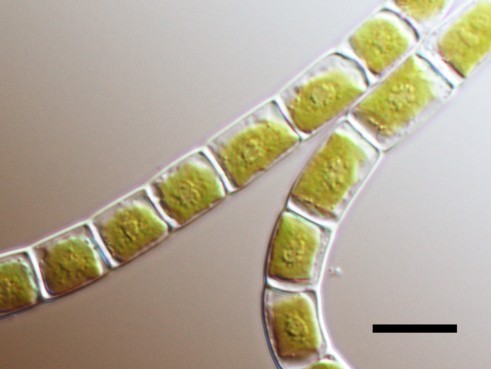
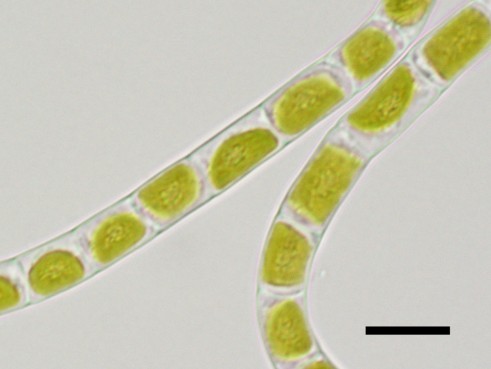
Left : microscope image (differential interference, X1000)
Right: microscope image (X1000)
Scale bars = 10 μm
Klebsormidium nitens NIES-2285 (formerly identified as K. flaccidum) was provided from the National Institute for Environmental Studies (NIES) of Japan.
Genome assemble status
- version 1.0 (Latest version) Reads
| Roche 454 GS FLX Titanium | Single run | 4,103,759 | reads |
| 3 Kbp mate pair | 1,483,727 | reads | |
| Illumina Genome Analyzer IIx | 75 bp paire | 54,525,596 | reads |
Statistics for assembly of the nuclear genome
| Number of scaffolds | 1,812 | scaffolds |
| Total scaffolds length | 103,921,766 | bp |
| Scaffold N50 size (N50) | 134,930 | bp (229 scaffolds) |
| Contig N50 size | 56,148 | bp |
| Peak coverage | 40 |
Organellar genomes status
- version 1.0 (Latest version) Statistical analysis of organellar genomes
| Chloroplast | genome size | 181,482 | bp |
| (scaffold kfl01813) | Protein-coding genes | 117 | genes |
| tRNA | 36 | genes | |
| rRNA | 6 | genes | |
| Mitochondrion | genome size | 106,468 | bp |
| (scaffold kfl01814) | Protein-coding genes | 35 | genes |
| tRNA | 29 | genes | |
| rRNA | 3 | genes |
Predicted transcripts status
- version 1.1 (Latest version) Statistical analysis of transcripts
- version 1.0 (Old version) Statistical analysis of transcripts
| Roche 454 GS FLX Titanium | Single run | 1,457,422 | reads |
| Illumina GAIIx | Single 69 bp | 215,698,946 | reads |
Statistical analysis of nuclear genes
| Predicted protein-coding genes | 17,055 | genes |
| Genes predicted both 5' UTR and 3' UTR | 9,100 | genes |
| Genes predicted 5' UTR only | 780 | genes |
| Genes predicted 3' UTR only | 1,353 | genes |
| Roche 454 GS FLX Titanium | Single run | 1,457,422 | reads |
| 18,661 | Isotigs | ||
| 17,422 | Isogroups |
Statistical analysis of nuclear genes
| Predicted protein-coding genes | 16,063 | genes |
| At least partially supported by assembled transcripts | 10,731 | genes (66.2%) |
| Fully supported by assembled transcripts | 7,376 | genes (45.5%) |
Publications
- 1) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation K Hori, F Maruyama, T Fujisawa, T Togashi, N Yamamoto, M Seo, S Sato, T Yamada, H Mori, N Tajima, T Moriyama, M Ikeuchi, M Watanabe, H Wada, K Kobayashi, M Saito, T Masuda, Y Sasaki-Sekimoto, K Mashiguchi, K Awai, M Shimojima, S Masuda, M Iwai, T Nobusawa, T Narise, S Kondo, H Saito, R Sato, M Murakawa, Y Ihara, Y Oshima-Yamada, K O, M Satoh, K Sonobe, M Ishii, R Ohtani, M Kanamori-Sato, R Honoki, D Miyazaki, H Mochizuki, J Umetsu, K Higashi, D Shibata, Y Kamiya, N Sato, Y Nakamura, S Tabata, S Ida, K Kurokawa, H Ohta
Nature communications 5, 4978 (doi:10.1038/ncomms4978)
(Klebsormidium flaccidum NIES-2285 was re-identified as Klebsormidium nitens NIES-2285, 2016 Jun)
